
BIOCHEMISTRY AND MOLECULAR BIOLOGY DEPARTMENT
In this issue of the newsletter, we will be highlighting the research being carried out by the Biochemistry and Molecular Biology Department at DDI, led by Dr. Jehad Abubaker.
MEET THE SCIENTIST
Meet the scientists behind DDI’s research
Epigenetics Applications In Type 2 Diabetes: Rna Methylation Can Regulate Human SS-cell Biology And Insulin Secretion
Published on 01/09/2019
The regulation of islet cell biology is critical for glucose homeostasis. Multiple studies demonstrated that m6A mediators (writers, erasers and readers) are expressed in human islets from control groups. However, analysis of T2D human islet cells showed that several m6A modulators (e.g. METTL3, ALKBH5, and YTHDF1) are downregulated in whole islets of T2D samples compared to their controls. Further analysis revealed that it is particularly β-cells that express decreased levels of m6A mediators due to T2D.
A recent study by De Jesus et. al (De Jesus et al., 2019) further investigated this observation and deciphered the mechanism by which RNA methylation could be affected in T2D. They assessed the differences in gene expression at the single-cell level specifically in β-cells in islets from controls and humans with T2D. They showed that m6A genes presented significantly higher variability compared to non-m6A genes, and within the m6A genes the coefficient of variation was higher in individuals with T2D than in controls. Interestingly, they showed that the genes affected in T2D are involved in cell-cycle regulation, receptor signaling, insulin secretion and pancreas development. Furthermore, these “hypomethylated” genes are within several pathways that are important for β-cell biology and are associated with the development of diabetes. Examples of these genes include maturity-onset diabetes of the young (MODY) and insulin receptor (IR)/insulin-like growth factor 1 receptor (IGF1R) signaling. More specifically, the insulin/IGF1–AKT–PDX1 pathway was affected significantly by hypomethylation in T2D. In fact, it is well-known that impairment of insulin/IGF1–AKT signaling followed by downregulation of PDX1 is associated with T2D. Together, their data suggest that hypomethylation of specific transcripts seen in T2D islets leads to cell-cycle arrest and impaired insulin secretion through downregulation of the insulin/IGF1–AKT–PDX1 pathway (Figure 5).
They were able to confirm their findings by using a human β-cell line and knocking down key m6A regulators (METTL3 or METTL14) then performing m6A and RNA sequencing. Furthermore, they generated Mettl14 β-cell-specific knockout mice (M14KO) to validate their findings in an in vivo mammalian system. They demonstrated that this knockdown can change the expression and methylation of transcripts involved in cell-cycle regulation and insulin secretion (e.g. PDX1 and IGF1R).
Finally, the study proposes that therapeutic targeting of m6A regulators in a β-cell-specific manner might be a new opportunity to counter the decreased m6A levels in T2D islets and to promote β-cell survival and function.
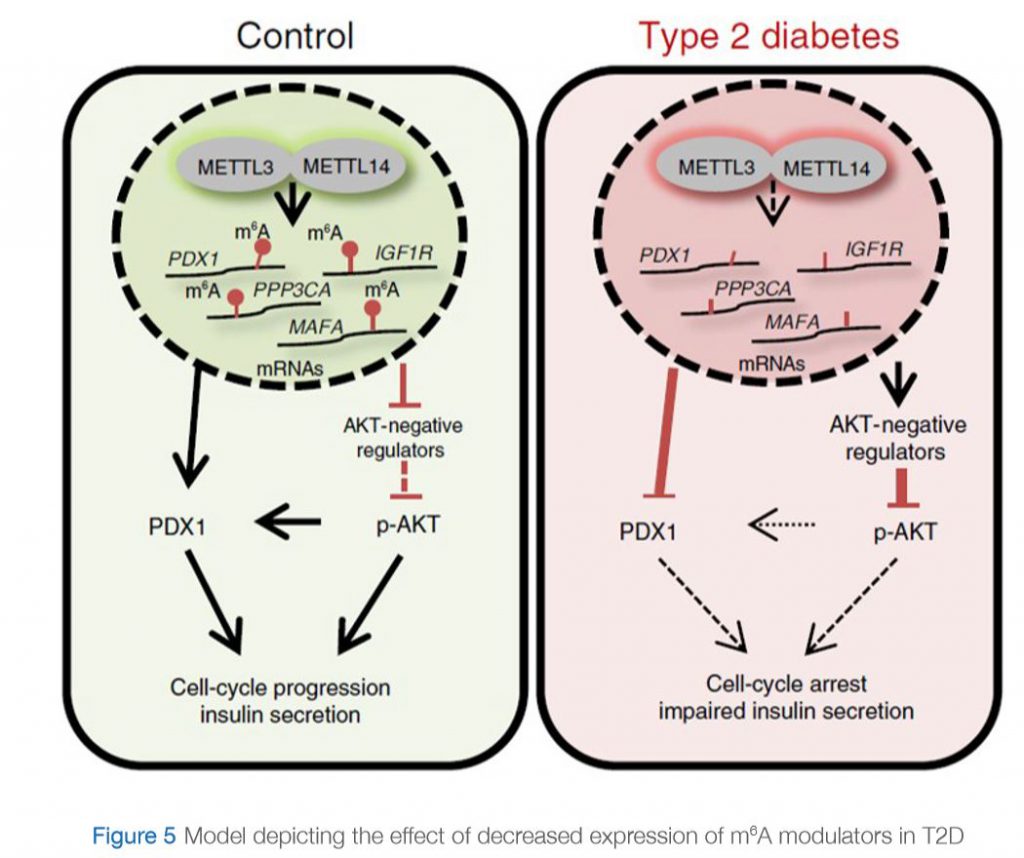
IN CONTROL STATE: Expression of “writers” (METTL3 and METTL14) is normal. Genes involved in insulin signaling are properly methylated. pAKT pathway is activated and PDX1 protein is produced leading to cell cycle progression and insulin secretion.
IN T2D: Expression of “writers” (METTL3 and METTL14) is reduced. Genes involved in insulin signaling are hypomethylated (lack methylation). This results in an increase in expression of negative regulators of AKT, leading to decreased phosphorylation of AKT. PDX1 expression is downregulated. This eventually leads to cell-cycle arrest and impaired insulin secretion.
REFERENCES:
1. Abu-Farha, M., P. Cherian, I. Al-Khairi, R. Nizam, A. Alkandari, H. Arefanian, J. Tuomilehto, F. Al-Mulla and J. Abubaker (2019). “Reduced miR-181d level in obesity and its role in lipid metabolism via regulation of ANGPTL3.” Scientific Reports 9(1): 11866.
2. De Jesus, D. F., Z. Zhang, S. Kahraman, N. K. Brown, M. Chen, J. Hu, M. K. Gupta, C. He and R. N. Kulkarni (2019). “m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes.” Nature Metabolism 1(8): 765-774.
3. Dominissini, D. (2014). “Roadmap to the epitranscriptome.” Science 346(6214): 1192.
4. Yang, Y., P. J. Hsu, Y. S. Chen and Y. G. Yang (2018). “Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism.” Cell Res 28(6): 616-624.


